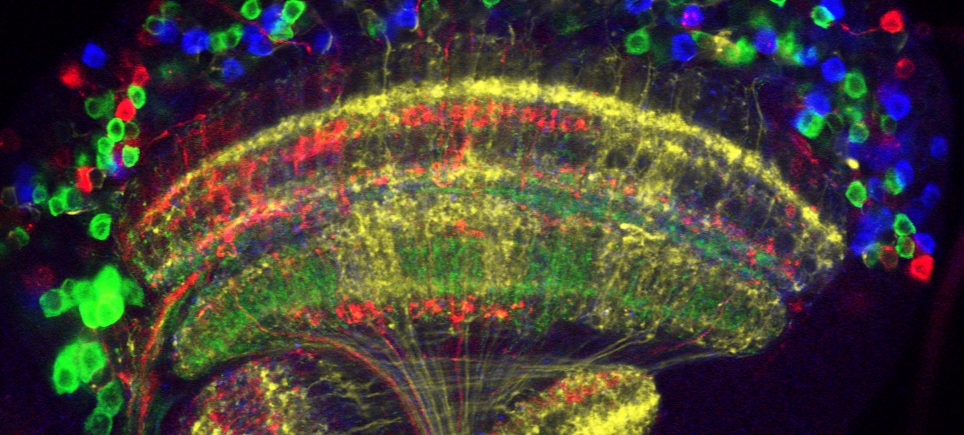
Inspired by the pioneering mouse Brainbow strategies of Livet et al. (Nature, 2007), we developed the Flybow multicolor labeling tool set for Drosophila. To take advantage of the increasing number of available Gal4 lines for controlling expression in any genetically accessible cell subpopulation and tissue of interest, Flybow relies on UAS-activated transcription. Furthermore, Flybow transgenes use the modified mFLP5–mFRT71 system which can be combined with the canonical FLP–FRT system. Their combination enables sparse labeling and if used as intersectional tools helps to refine expression. Strengths of the Flybow approach are that it enables direct imaging of endogenous fluorescence signals in four channels and only one UAS transgene is required.
Flybow-1.0 and –1.1 constructs consist of one or two invertible cassettes, mediating the stochastic expression of two or four fluorescent proteins, respectively. In Flybow-2.0, an additional transcriptional stop cassette flanked by FRT sites in the same orientation precedes the cassettes to eliminate default marker expression. The stop cassette is excised after induction of the canonical FLP recombinase.
The original set of Flybow constructs uses an epitope-tagged cyan fluorescent protein mCerulean variant, which requires immunodetection because of its weak native emission in flies. In a second set of transgenes (Flybow-1.0B, 1.1B and 2.0B), mCerulean-V5 was replaced by the brighter mTurquoise. A third Flybow family (Flybow-1.0C, 1.1C and 2.0C) uses TagRFP-T and mTurquoise2.

Further information about Flybow transgenes and how they are used practically can be found in below publications:
Hadjieconomou, D., Rotkopf, S., Alexandre, C., Bell, D.M., Dickson, B.J. and Salecker, I. (2011) Flybow: genetic multicolor cell-labeling for neural circuit analysis in Drosophila melanogaster. Nature Methods 8, 260-266.
doi.org/10.1038/nmeth.1567
Shimosako, N., Hadjieconomou, D., and Salecker, I. (2014) Flybow to dissect circuit assembly in the Drosophila brain. Methods Mol. Biol. 1082, 57-69. – book chapter, includ. original data.
doi.org/10.1007/978-1-62703-655-9_4
Richier, B. and Salecker, I. (2015) Versatile genetic paintbrushes: Brainbow technologies. WIREs Developmental Biology 4, 161-180. – Review.
doi.org/10.1002/wdev.166
Apitz, H., and Salecker, I. (2018) Spatio-temporal relays control layer identity of direction-selective neuron subtypes in Drosophila. Nature Communications 9, 2295.
doi.org/10.1038/s41467-018-04592-z
Powell, E.L. and Salecker, I. (2019/2020) Flybow to dissect circuit assembly in the Drosophila brain: an update. Methods Mol. Biol. 2047, 137-152.
doi.org/10.1007/978-1-4939-9732-9_8
Flybow and Flybow B family, as well as hs-mFLP5 stocks can be obtained from the Drosophila Bloomington Stock Center (BDSC). For plasmids, maps, Flybow C and other stocks, please contact Iris.